MGEScan on Galaxy
A Galaxy based system for identifying retrotransposons in genome
A computational method MGEScan is now available on Galaxy Workflow System with various computing environments including Amazon Cloud. With Galaxy tools for MGEScan, each step of MGEScan are described in workflows and visualized its output through UCSC Genome Browser or Ensembl Genome Browser. Amazon EC2 allows you to create a server for MGEScan on Galaxy within 5 minutes including all required tools such as EMBOSS, HMMER 3.0+, RepeatMasker, and Tandem Repeats Finder (trf). The installable package on github enables you to have a private server for MGEScan with Galaxy Workflow System. Having a private server of MGEScan is useful if there are security concerns of data analysis or personalized tools. MGESCan is also available via Galaxy Tool Shed and the public server (usegalaxy.org) as a published workflow.
Sample Datasets are available on the demo site at Indiana University. It gives you an overview of using MGEScan on Galaxy Workflow System.
| Demo (indiana.edu) |
|---|
 |
MGEScan on Galaxy is accessible via four choices: Amazon Cloud, Local Download, Galaxy Tool Shed, or Galaxy Published Workflow.
| On Amazon Cloud EC2 | Download (local) |
|---|---|
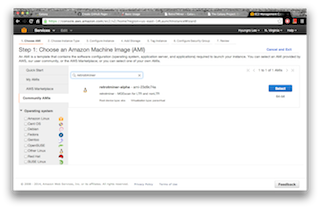 |
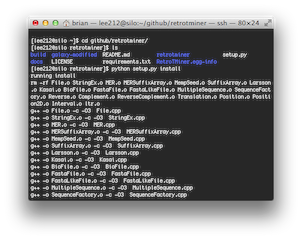 |
| Galaxy Tool Shed | Published MGEScan Workflow |
|---|---|
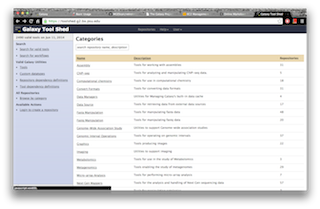 |
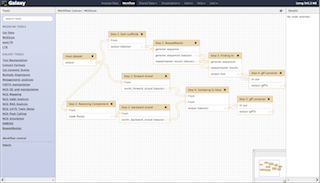 |
DEMO Version : silo.cs.indiana.edu